Abstract
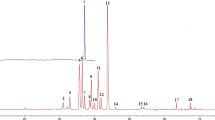
Chromatographic fingerprinting is an effective methodology for authentication and quality control of herbal products. In the presented study, a chemometric strategy based on multivariate curve resolution–alternating least squares (MCR–ALS) and multivariate pattern recognition methods was used to establish a gas chromatography–mass spectrometry (GC–MS) fingerprint of saffron. For this purpose, the volatile metabolites of 17 Iranian saffron samples, collected from different geographical regions, were determined using the combined method of ultrasound-assisted solvent extraction (UASE) and dispersive liquid–liquid microextraction (DLLME), coupled with GC–MS. The resolved elution profiles and the related mass spectra obtained by an extended MCR–ALS algorithm were then used to estimate the relative concentrations and to identify the saffron volatile metabolites, respectively. Consequently, 77 compounds with high reversed match factors (RMFs > 850) were successfully determined. The relative concentrations of these compounds were used to generate a new data set which was analyzed by multivariate data analysis methods including principal component analysis (PCA) and k-means. Accordingly, the saffron samples were categorized into five classes using these techniques. The results revealed that 11 compounds, as biomarkers of saffron, contributed to the class discrimination and characterization. Eleven biomarkers including nine secondary metabolites of saffron (safranal, α- and β-isophorone, phenylethyl alcohol, ketoisophorone, 2,2,6-trimethyl-1,4-cyclohexanedione, 2,6,6-trimethyl-4-oxo-2-cyclohexen-1-carbaldehyde, 2,4,4-trimethyl-3-carboxaldehyde-5-hydroxy-2,5-cyclohexadien-1-one, and 2,6,6-trimethyl-4-hydroxy-1-cyclohexene-1-carboxaldehyde (HTCC)), a primary metabolite (linoleic acid), and a long chain fatty alcohol (nanocosanol) were distinguished as the saffron fingerprint. Finally, the individual contribution of each biomarker to the classes was determined by the counter propagation artificial neural network (CPANN) method.

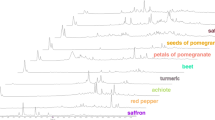
General framework of the purposed pattern recognition method for saffron






Similar content being viewed by others
References
Lim TK. Crocus sativus. In: Edible medicinal and non-medicinal plants, vol. 8. Dordrecht: Springer Netherlands; 2014. p. 77–136.
Melnyk JP, Wang S, Marcone MF. Chemical and biological properties of the world's most expensive spice: saffron. Food Res Int. 2010;43(8):1981–9.
Fernández J-A. Anticancer properties of saffron, Crocus sativus Linn. In: Mahmud THK, Arjumand A, editors. Advances in phytomedicine, vol. 2. Amsterdam: Elsevier; 2006. p. 313–30.
Hosseinzadeh H. Saffron: a herbal medicine of third millennium. Jundishapur J Nat Pharm Prod. 2014;9(1):1–2.
Jalali-Heravi M, Parastar H, Ebrahimi-Najafabadi H. Characterization of volatile components of Iranian saffron using factorial-based response surface modeling of ultrasonic extraction combined with gas chromatography–mass spectrometry analysis. J Chromatogr A. 2009;1216(33):6088–97.
Anastasaki E, Kanakis C, Pappas C, Maggi L, del Campo CP, Carmona M, et al. Geographical differentiation of saffron by GC–MS/FID and chemometrics. Eur Food Res Technol. 2009;229(6):899–905.
Jalali-Heravi M, Parastar H, Ebrahimi-Najafabadi H. Self-modeling curve resolution techniques applied to comparative analysis of volatile components of Iranian saffron from different regions. Anal Chim Acta. 2010;662(2):143–54.
Anastasaki E, Kanakis C, Pappas C, Maggi L, del Campo CP, Carmona M, et al. Differentiation of saffron from four countries by mid-infrared spectroscopy and multivariate analysis. Eur Food Res Technol. 2010;230(4):571–7.
Maggi L, Carmona M, Kelly SD, Marigheto N, Alonso GL. Geographical origin differentiation of saffron spice (Crocus sativus L. stigmas) – preliminary investigation using chemical and multi-element (H, C, N) stable isotope analysis. Food Chem. 2011;128(2):543–8.
Ordoudi SA, de los Mozos Pascual M, Tsimidou MZ. On the quality control of traded saffron by means of transmission Fourier-transform mid-infrared (FT-MIR) spectroscopy and chemometrics. Food Chem. 2014;150:414–21.
Maggi L, Carmona M, Zalacain A, Kanakis CD, Anastasaki E, Tarantilis PA, et al. Changes in saffron volatile profile according to its storage time. Food Res Int. 2010;43(5):1329–34.
Gresta F, Lombardo GM, Siracusa L, Ruberto G. Saffron, an alternative crop for sustainable agricultural systems: a review. In: Lichtfouse E, Navarrete M, Debaeke P, Véronique S, Alberola C, editors. Sustainable agriculture. Dordrecht: Springer Netherlands; 2009. p. 355–76.
Yilmaz A, Nyberg N, Mølgaard P, Asili J, Jaroszewski J. 1H NMR metabolic fingerprinting of saffron extracts. Metabolomics. 2010;6(4):511–7.
Petrakis EA, Cagliani LR, Polissiou MG, Consonni R. Evaluation of saffron (Crocus sativus L.) adulteration with plant adulterants by 1H NMR metabolite fingerprinting. Food Chem. 2015;173:890–6.
Goodarzi M, Russell PJ, Vander Heyden Y. Similarity analyses of chromatographic herbal fingerprints: a review. Anal Chim Acta. 2013;804:16–28.
Hakimzadeh N, Parastar H, Fattahi M. Combination of multivariate curve resolution and multivariate classification techniques for comprehensive high-performance liquid chromatography-diode array absorbance detection fingerprints analysis of Salvia reuterana extracts. J Chromatogr A. 2014;1326:63–72.
Tistaert C, Dejaegher B, Heyden YV. Chromatographic separation techniques and data handling methods for herbal fingerprints: a review. Anal Chim Acta. 2011;690(2):148–61.
Sereshti H, Heidari R, Samadi S. Determination of volatile components of saffron by optimised ultrasound-assisted extraction in tandem with dispersive liquid–liquid microextraction followed by gas chromatography–mass spectrometry. Food Chem. 2014;143:499–505.
Kanakis CD, Daferera DJ, Tarantilis PA, Polissiou MG. Qualitative determination of volatile compounds and quantitative evaluation of safranal and 4-hydroxy-2,6,6-trimethyl-1-cyclohexene-1-carboxaldehyde (HTCC) in Greek saffron. J Agric Food Chem. 2004;52(14):4515–21.
D'Auria M, Mauriello G, Rana GL. Volatile organic compounds from saffron. Flavour Fragrance J. 2004;19(1):17–23.
Hendriks MMWB, Eeuwijk FAV, Jellema RH, Westerhuis JA, Reijmers TH, Hoefsloot HCJ, et al. Data-processing strategies for metabolomics studies. TrAC Trends Anal Chem. 2011;30(10):1685–98.
Liland KH. Multivariate methods in metabolomics – from pre-processing to dimension reduction and statistical analysis. TrAC Trends Anal Chem. 2011;30(6):827–41.
Parastar H, Tauler R. Multivariate curve resolution of hyphenated and multidimensional chromatographic measurements: a new insight to address current chromatographic challenges. Anal Chem. 2013;86(1):286–97.
Jalali-Heravi M, Parastar H. Recent trends in application of multivariate curve resolution approaches for improving gas chromatography–mass spectrometry analysis of essential oils. Talanta. 2011;85(2):835–49.
Bansal A, Chhabra V, Rawal RK, Sharma S. Chemometrics: a new scenario in herbal drug standardization. J Pharm Anal. 2014;4(4):223–33.
Parastar H, Jalali-Heravi M, Sereshti H, Mani-Varnosfaderani A. Chromatographic fingerprint analysis of secondary metabolites in citrus fruits peels using gas chromatography–mass spectrometry combined with advanced chemometric methods. J Chromatogr A. 2012;1251:176–87.
Jaumot J, Gargallo R, de Juan A, Tauler R. A graphical user-friendly interface for MCR–ALS: a new tool for multivariate curve resolution in MATLAB. Chemom Intell Lab Syst. 2005;76(1):101–10.
Ballabio D, Vasighi M. A MATLAB toolbox for self organizing maps and supervised neural network learning strategies. Chemom Intell Lab Syst. 2012;118:24–32.
Jalali-Heravi M, Parastar H, Kamalzadeh M, Tauler R, Jaumot J. MCRC software: a tool for chemometric analysis of two-way chromatographic data. Chemom Intell Lab Syst. 2010;104(2):155–71.
Esbensen KH, Geladi P. Principal component analysis: concept, geometrical interpretation, mathematical background, algorithms, history, practice. In: Brown SD, Tauler R, Walczak B, editors. Comprehensive chemometrics, vol. 2. Oxford: Elsevier; 2009. p. 211–26.
Maeder M, de Juan A. Two-way data analysis: evolving factor analysis. In: Walczak SDBT, editor. Comprehensive chemometrics. Oxford: Elsevier; 2009. p. 261–74.
Windig W, Guilment J. Interactive self-modeling mixture analysis. Anal Chem. 1991;63(14):1425–32.
Tauler R, Maeder M, de Juan A. Multiset data analysis: extended multivariate curve resolution. In: Brown SD, Tauler R, Walczak B, editors. Comprehensive chemometrics, vol. 2. Oxford: Elsevier; 2009. p. 473–505.
Gong F, Liang Y-Z, Cui H, Chau F-T, Chan BT-P. Determination of volatile components in peptic powder by gas chromatography–mass spectrometry and chemometric resolution. J Chromatogr A. 2001;909(2):237–47.
Bro R, Smilde AK. Principal component analysis. Anal Methods. 2014;6(9):2812–31.
Sun L-X, Xu F, Liang Y-Z, Xie Y-L, Yu R-Q. Cluster analysis by the k-means algorithm and simulated annealing. Chemom Intell Lab Syst. 1994;25(1):51–60.
Zupan J, Novič M, Ruisánchez I. Kohonen and counterpropagation artificial neural networks in analytical chemistry. Chemom Intell Lab Syst. 1997;38(1):1–23.
Fidencio PH, Ruisanchez I, Poppi RJ. Application of artificial neural networks to the classification of soils from Sao Paulo State using near-infrared spectroscopy. Analyst. 2001;126(12):2194–200.
De Maesschalck R, Jouan-Rimbaud D, Massart DL. The Mahalanobis distance. Chemom Intell Lab Syst. 2000;50(1):1–18.
Acknowledgments
The study was carried out as a part of the research project titled “Development of an index for evaluation of Iranian saffron quality based on chemical constituents in its chromatographic fingerprints: combination of gas chromatograpy and novel multivariate chemometric methods” supported by Iran national science foundation (INSF) (http://www.insf.org). The authors would like thank to Mr. Mohammad-Ali Shoshtari and Mr. Ali Mokhtarian for their great help in supplying saffron samples from different plantation sites in Iran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 374 kb)
Rights and permissions
About this article
Cite this article
Aliakbarzadeh, G., Sereshti, H. & Parastar, H. Pattern recognition analysis of chromatographic fingerprints of Crocus sativus L. secondary metabolites towards source identification and quality control. Anal Bioanal Chem 408, 3295–3307 (2016). https://doi.org/10.1007/s00216-016-9400-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9400-8